Questions
BIOL3612.MERGED.202610 Activity Questions from the Literature 2B: Preparing tRNAs and synthesizing protein
Single choice
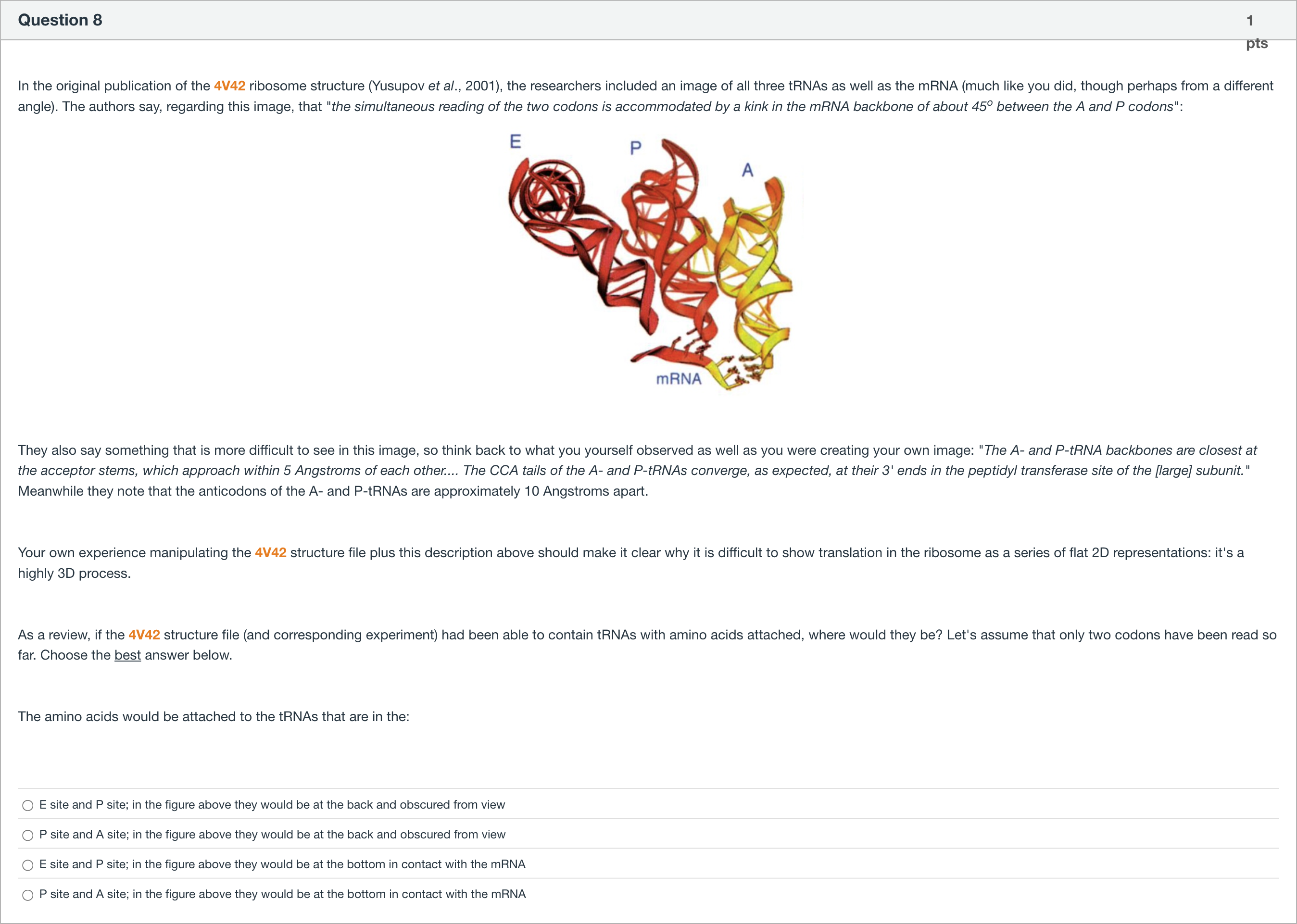
In the original publication of the 4V42 ribosome structure (Yusupov et al., 2001), the researchers included an image of all three tRNAs as well as the mRNA (much like you did, though perhaps from a different angle). The authors say, regarding this image, that "the simultaneous reading of the two codons is accommodated by a kink in the mRNA backbone of about 45o between the A and P codons": They also say something that is more difficult to see in this image, so think back to what you yourself observed as well as you were creating your own image: "The A- and P-tRNA backbones are closest at the acceptor stems, which approach within 5 Angstroms of each other.... The CCA tails of the A- and P-tRNAs converge, as expected, at their 3' ends in the peptidyl transferase site of the [large] subunit." Meanwhile they note that the anticodons of the A- and P-tRNAs are approximately 10 Angstroms apart. Your own experience manipulating the 4V42 structure file plus this description above should make it clear why it is difficult to show translation in the ribosome as a series of flat 2D representations: it's a highly 3D process. As a review, if the 4V42 structure file (and corresponding experiment) had been able to contain tRNAs with amino acids attached, where would they be? Let's assume that only two codons have been read so far. Choose the best answer below. The amino acids would be attached to the tRNAs that are in the:
Options
A.E site and P site; in the figure above they would be at the back and obscured from view
B.P site and A site; in the figure above they would be at the back and obscured from view
C.E site and P site; in the figure above they would be at the bottom in contact with the mRNA
D.P site and A site; in the figure above they would be at the bottom in contact with the mRNA
View Explanation
Verified Answer
Please login to view
Step-by-Step Analysis
To approach this question, I’ll break down what the scenario asks and then evaluate each answer in turn.
Option 1: "E site and P site; in the figure above they would be at the back and obscured from view". This option pairs E site with P site, but the scenario specifies two codons have been read and focuses on where the amino acids would be attached. The E site is where deacylated tRNAs exit, not where aminoacyl-tRNAs carrying amino acids would be located for peptidyl transfer. Additionally, describing their position as at the back and obscured from view doesn’t align with the described visibility o......Login to view full explanationLog in for full answers
We've collected over 50,000 authentic exam questions and detailed explanations from around the globe. Log in now and get instant access to the answers!
Similar Questions
Question at position 67 Translate the template DNA sequence into protein as a ribosome would. I recommend working this out on scrap paper, then finding the answer. Template DNA: 5'-TTTAAAACACCCACCGATGTGTCATT-3'N-Met-Cys-His-CN-Lys-Phe-Cys-Gly-Trp-His-His-Ser-CN-Met-Thr-His-Arg-Trp-Val-Phe-CNo protein is madeN-Met-Thr-His-Arg-Trp-Val-Phe-Trp-C
Question at position 64 Translate the coding DNA sequence into protein as a ribosome would. I recommend working this out on scrap paper, then finding the answer. Coding DNA: 5'-GCACTATGCCCCTGCGGGGATGAGTA-3'N-Met-Pro-Leu-Arg-Gly-CN-Ala-Leu-Cys-Pro-Cys-Gly-Asp-Glu-CN-Met-Pro-Leu-Arg-Gly-Trp-CNo protein is madeN-Met-Ser-Arg-Gly-Val-Pro-Val-C
Question at position 79 In both prokaryotes and eukaryotes, how many ribosomes can translate an mRNA at a time?twothreemanyone
A gene in the DNA Template Strand sequence is provided below. 3′ −G-G-A-A-C-G-T-A-C-T-A-A-C-G-G-A-C-T-A-T−5′ Given this sequence, what is the resulting amino acid sequence of the polypeptide following translation at the ribosome?
More Practical Tools for Students Powered by AI Study Helper
Making Your Study Simpler
Join us and instantly unlock extensive past papers & exclusive solutions to get a head start on your studies!